Python Simulations of Chemistry Framework
Outline
- What is PySCF?
- Feature Highlights
- How does PySCF connect to other (CCQ) projects?
- Installation
- Open-source community and documentation
What does PySCF do?
Mostly generic two-body Hamiltonians: \[ \hat{\text{H}} = \sum_{pq} h_{pq} \hat{a}_p^\dagger \hat{a}_q + \sum_{pqrs} V_{pqrs} \hat{a}_p^\dagger \hat{a}_q \hat{a}_r^\dagger \hat{a}_s \]Special forms (e.g. sparsity of V) are rarely exploited.
PySCF calculates \(h_{pq} \) and \( V_{pqrs} \) or users can provide them
Several basis options: Gaussian-type orbitals (GTOs) for molecules and solids and plane-wave
What does PySCF do (pt. 2)?
- Energies (ground and excited state)
- Forces and geometry optimization
- Response properties (e.g. dipole moments)
- Growing support for spectroscopic properties
- Relativstic effects and non-collinear spins
- Users can study systems at many levels of theory
Design Philosophy
A sandbox, but a performant one
Python user interface with a C
back-end
-------------|----------------------------------------------------
Language | files blank comment code
-------------|----------------------------------------------------
Python | 742 33356 42117 170260
C | 65 2888 2825 41259
Lisp | 5 78 145 886
C/C++ Header | 18 176 250 640
CMake | 12 68 203 268
Fortran 90 | 2 60 56 159
Bourne Shell | 5 20 25 81
-------------|----------------------------------------------------
SUM: | 851 36652 45628 213574
-------------|----------------------------------------------------
Correlated Methods
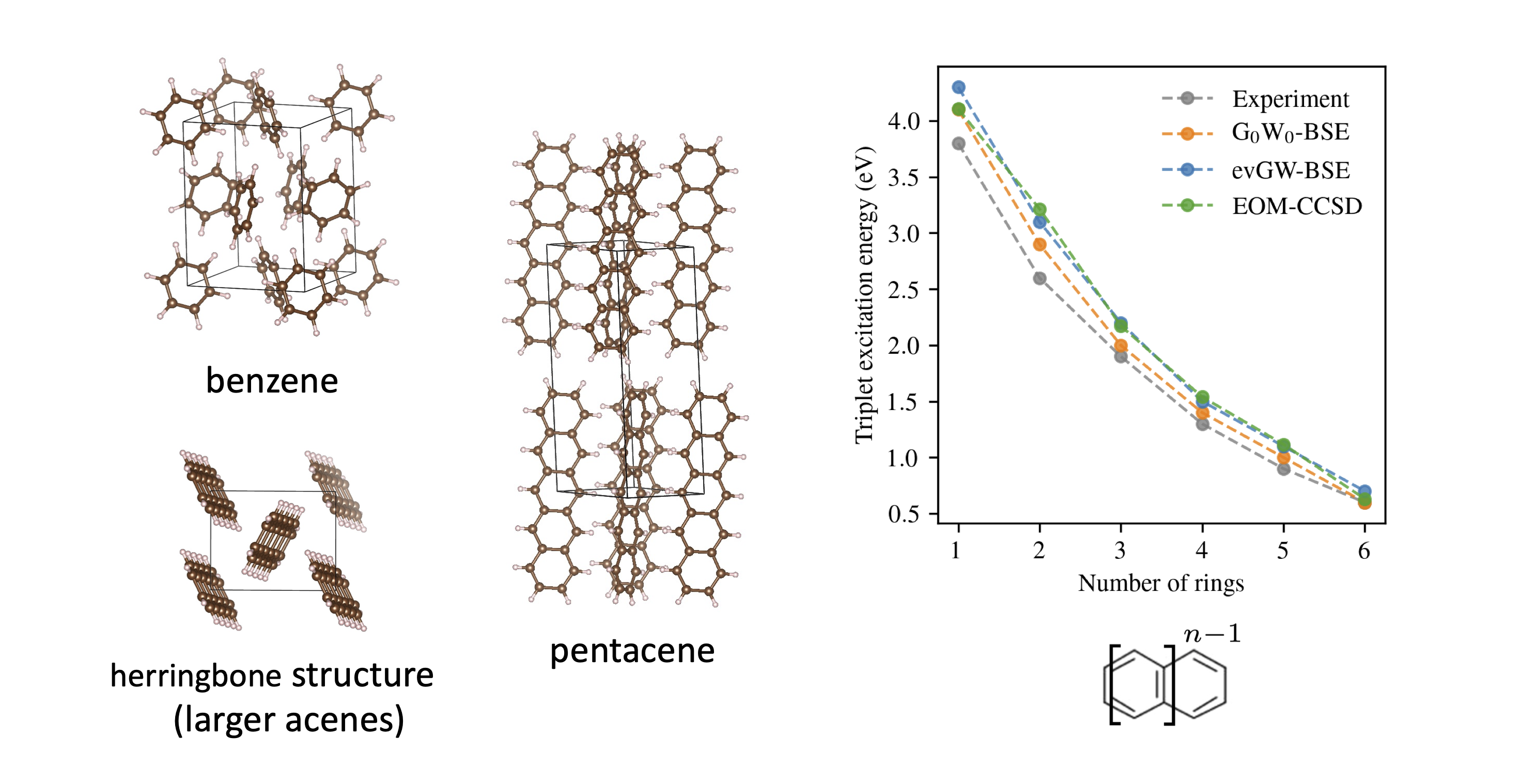
We have a lot of correlated methods that work well in molecules and solids
Xiao Wang
And now... plane waves
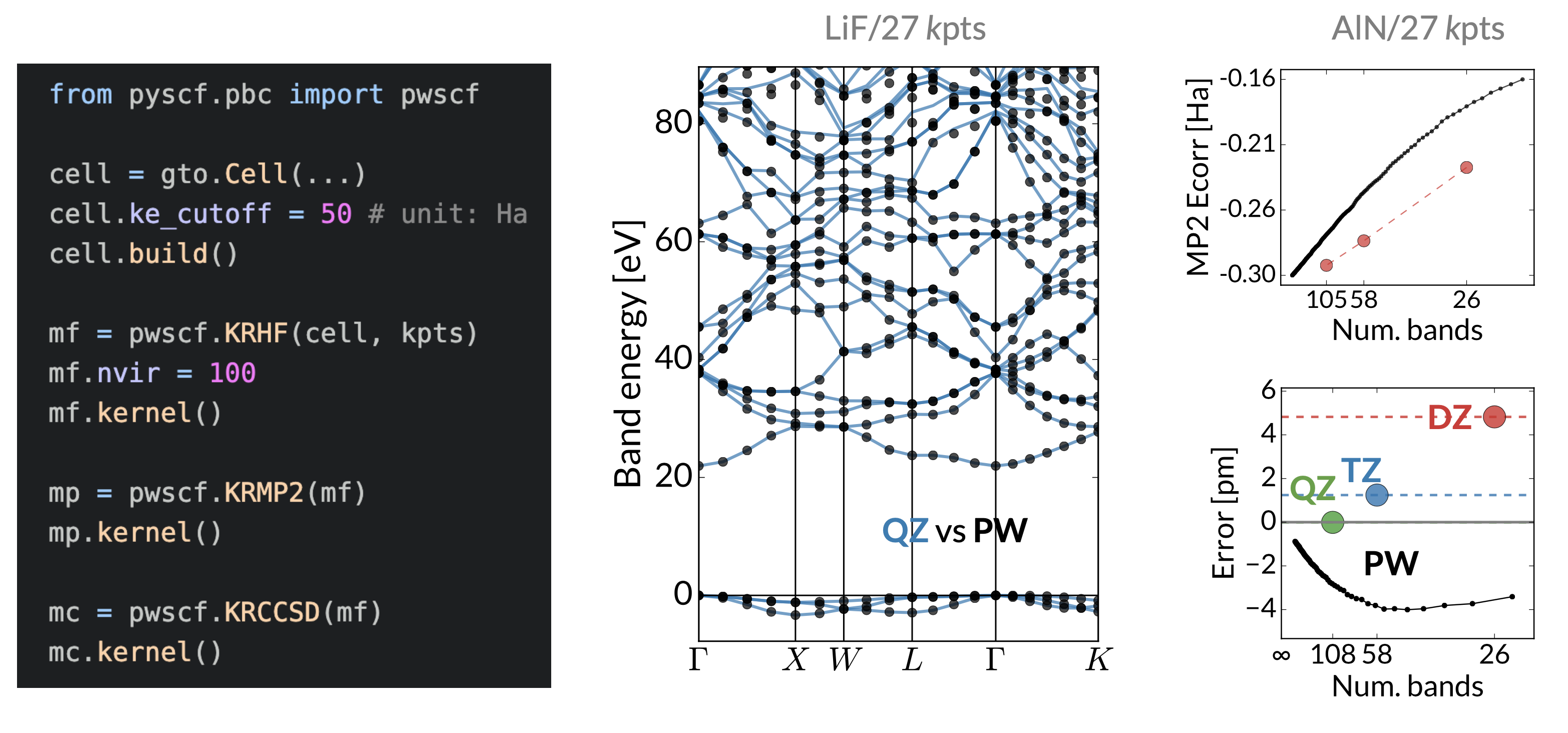
Hongzhou Ye and Verena Neufeld
Custom Hamiltonians
h1 = numpy.zeros((n,n))
for i in range(n-1):
h1[i,i+1] = h1[i+1,i] = -1.0
h1[n-1,0] = h1[0,n-1] = -1.0
eri = numpy.zeros((n,n,n,n))
for i in range(n):
eri[i,i,i,i] = 2.0
mf = scf.RHF(mol)
mf.get_hcore = lambda *args: h1
mf.get_ovlp = lambda *args: numpy.eye(n)
mf.kernel()
mymp = mp.MP2(mf)
mymp.kernel()
mycc = cc.CCSD(mf)
mycc.kernel()
mycas = mcscf.CASSCF(mf, 4, 4)
mycas.kernel()MPI4PySCF
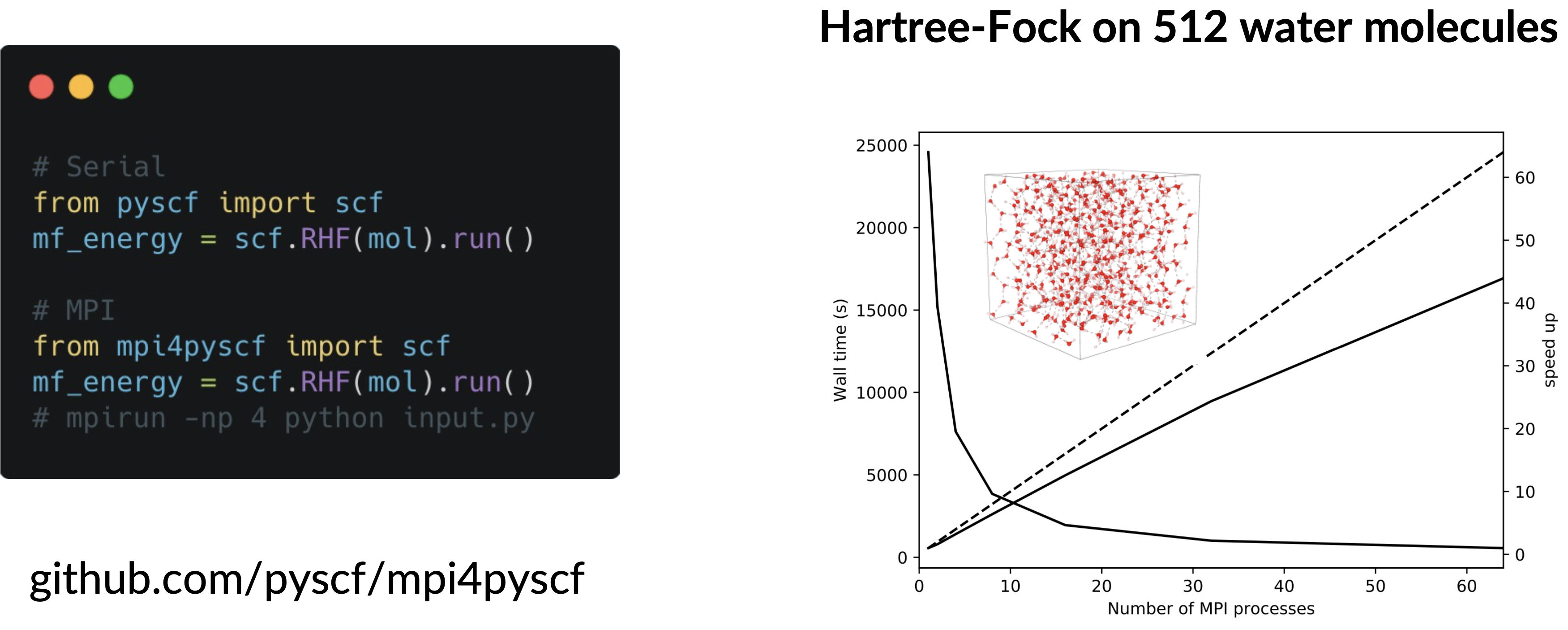
Interfaces with PySCF
DMRG
Block (Chan)
CheMPS2 (Wouters)
Block (Chan)
CheMPS2 (Wouters)
Selected CI
Arrow (Umrigar)
Dice (Sharma)
Arrow (Umrigar)
Dice (Sharma)
QMC
AFQMC (Morales/Zhang)
QMCPACK (LLNL)
NECI (Alavi)
Hande (Thom)
AFQMC (Morales/Zhang)
QMCPACK (LLNL)
NECI (Alavi)
Hande (Thom)
NN VMC
FermiNet (Deepmind)
DeepQMC/PauliNet (Noe)
NetKet (Carleo)
FermiNet (Deepmind)
DeepQMC/PauliNet (Noe)
NetKet (Carleo)
Quantum Computing
OpenFermion
Qiskit (IBM)
OpenQemist (1Qbit)
OpenFermion
Qiskit (IBM)
OpenQemist (1Qbit)
Somewhere in the future?
ITensor, TRIQS, your research code!
ITensor, TRIQS, your research code!
How can I get PySCF?
Option 1: Using pip
pip install pyscf
Option 2: Using pip to installing the latest version
pip install -e git+https://github.com/pyscf/pyscf.gitOption 3: Build PySCF from sources
Requirements:
- C and Fortran Compiler
- BLAS
- CMake
- Python 3, NumPy, SciPy, h5py
git clone https://github.com/pyscf/pyscf.git
cd pyscf/pyscf/lib
cmake -B build
cmake --build build --parallel
Documentation
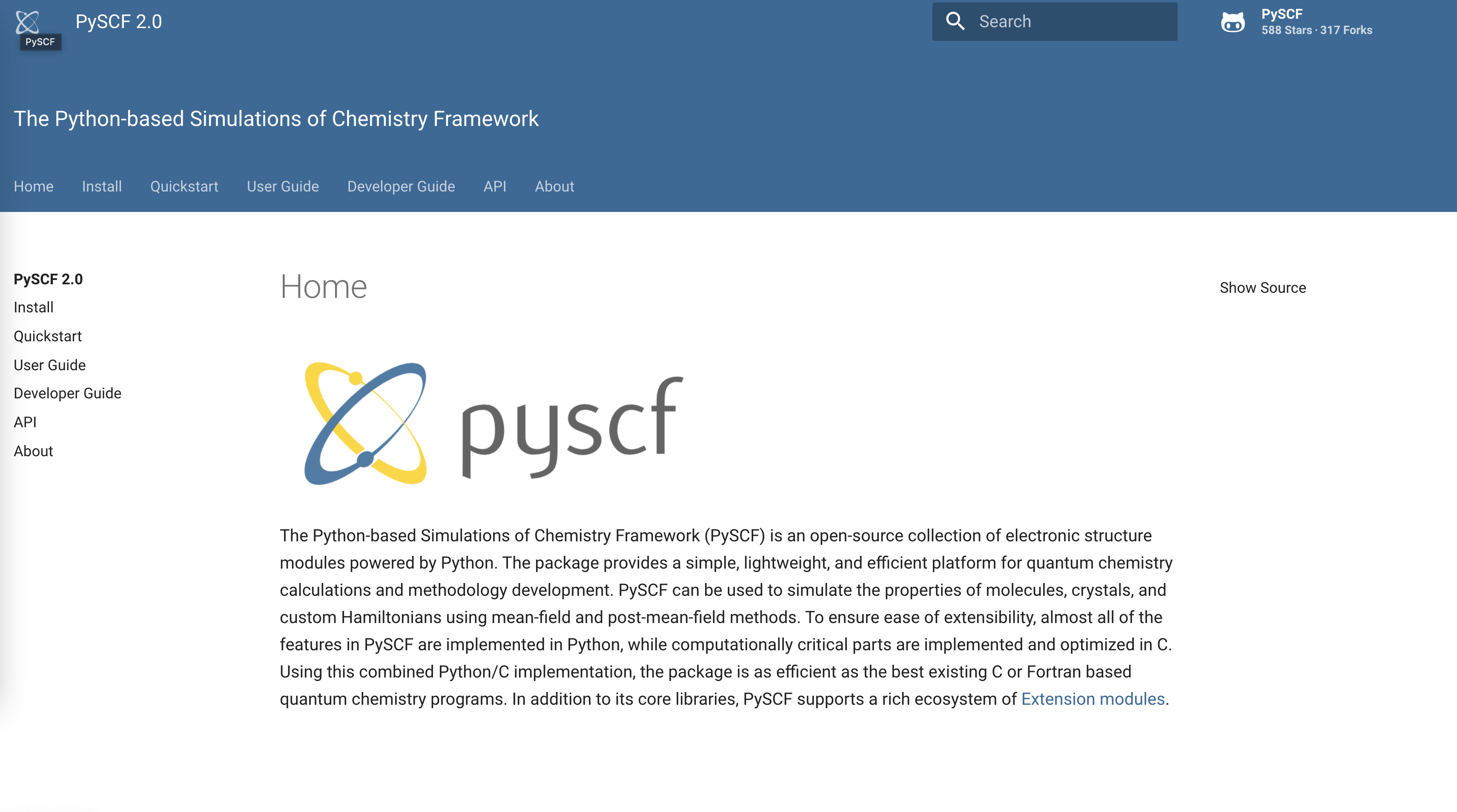
Documentation
Examples
Over 350 examples!!
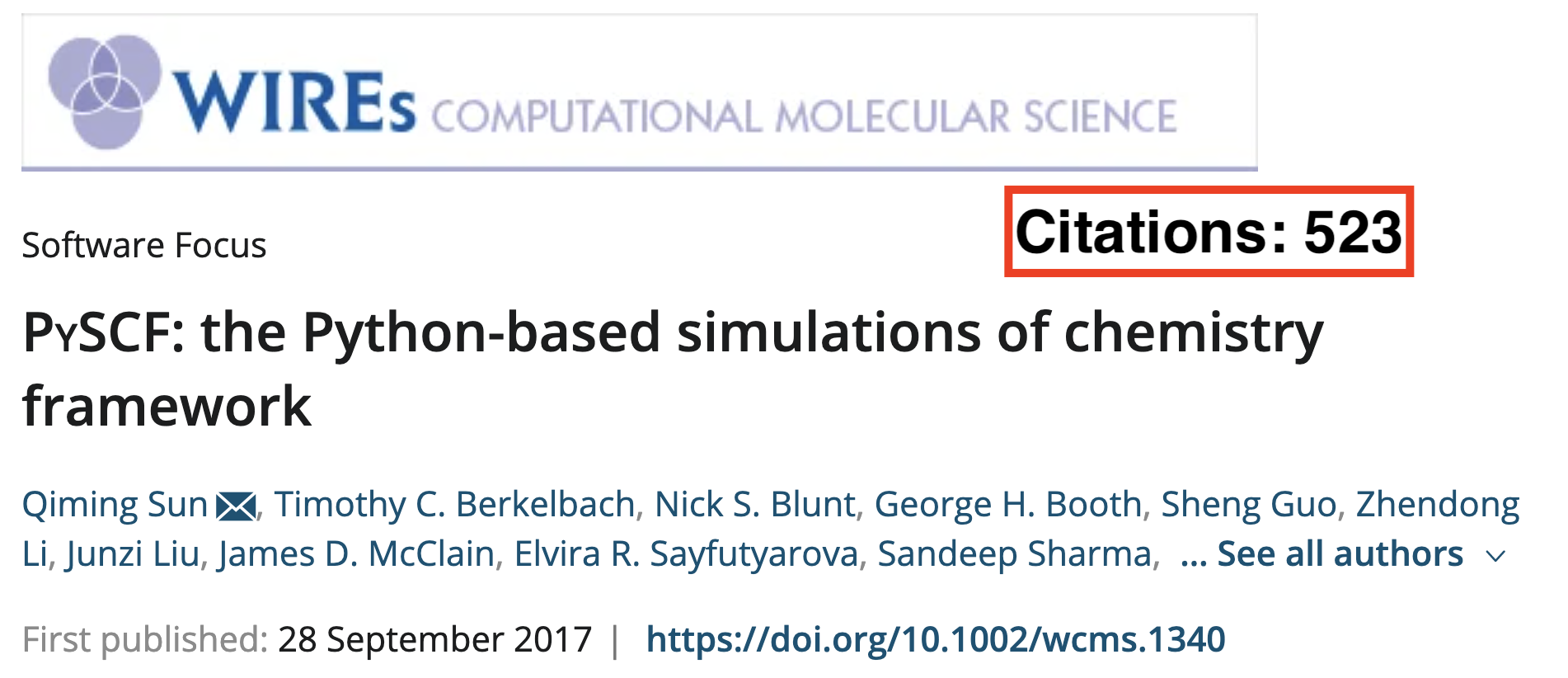
Open Source Community
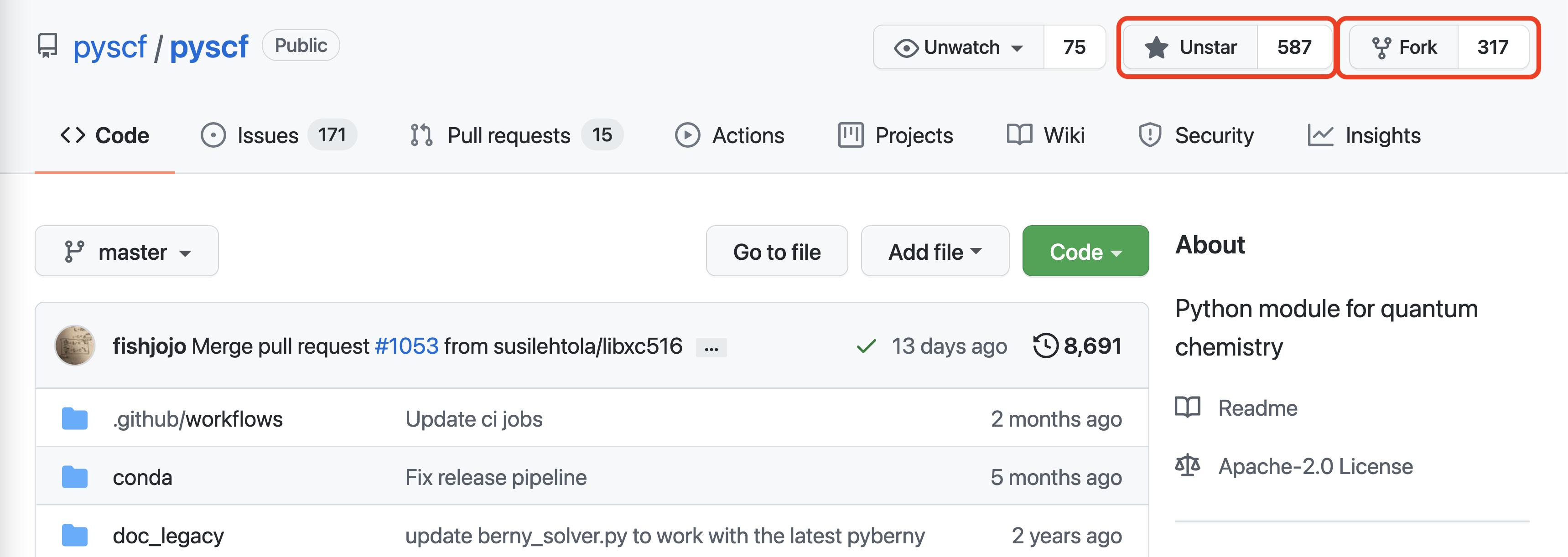
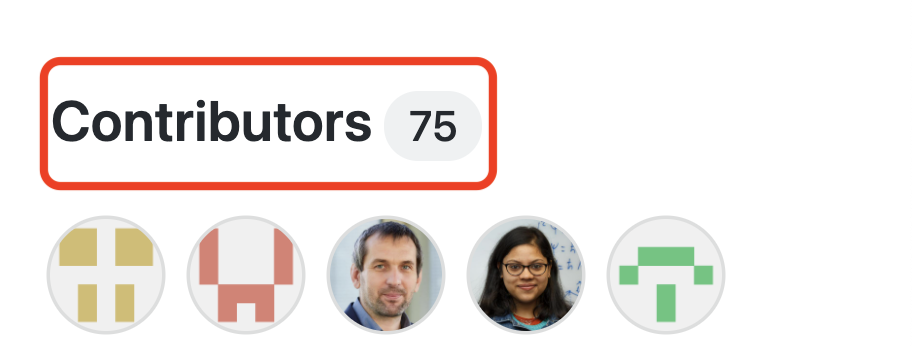
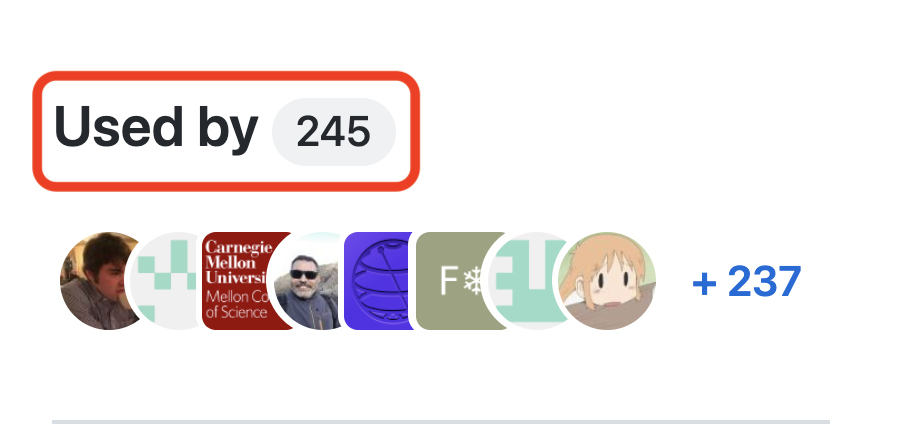
Impact

HF
MP2
CCSD
CASSCF
DFT
GW
TDDFT
EOM-CCSD
MRPT
ADC
FCI
X2C
MUCH MORE!
- Multiple levels of theory to calculate properties
- If you see something you want and it's not in PySCF already, talk to me!
- Check out the PySCF channel on the CCQ slack
- Let us know if you want to get involved!
- Hackathon for CCQ codes to accelerate interfaces?!?
EXTRA SLIDES
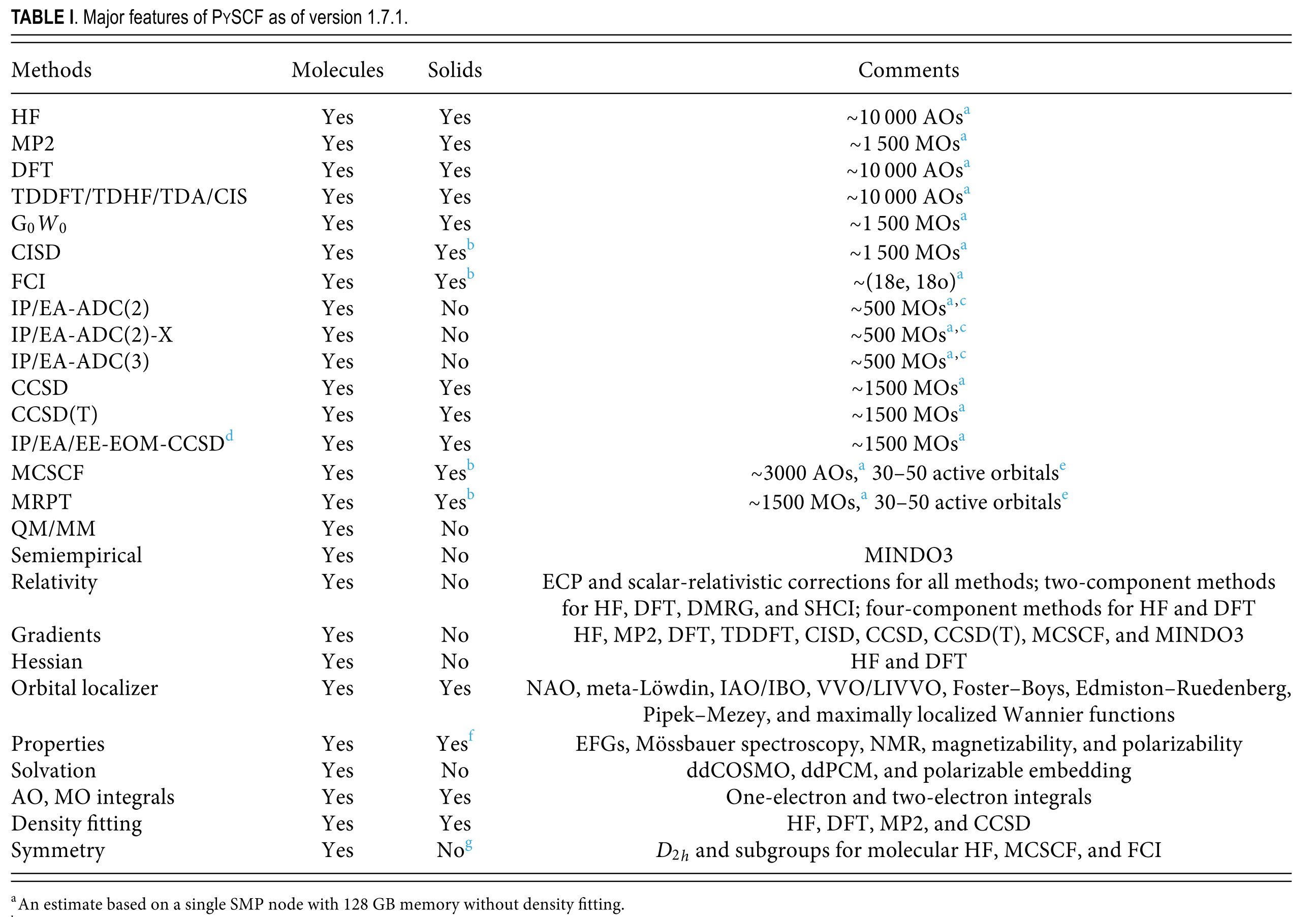
Capabilities in Detail